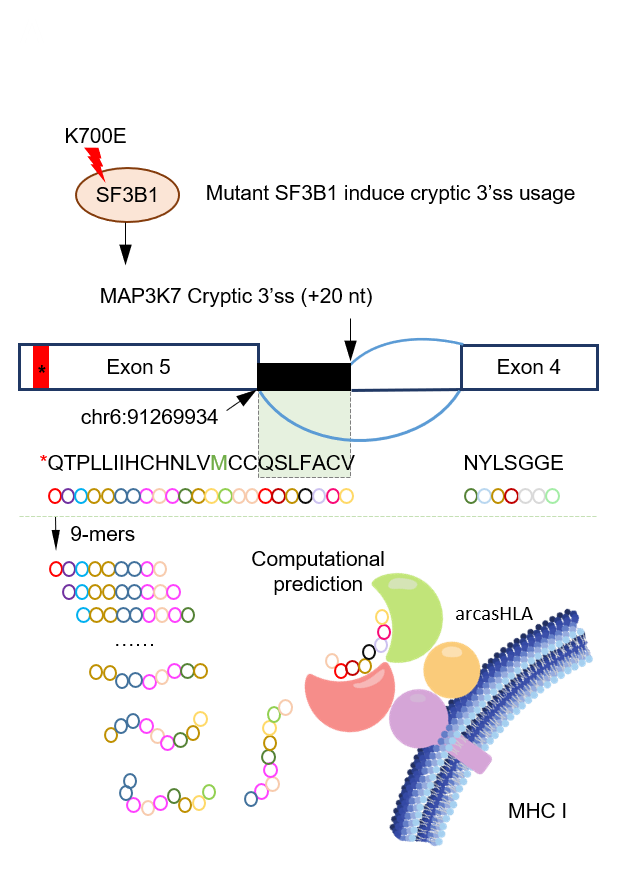
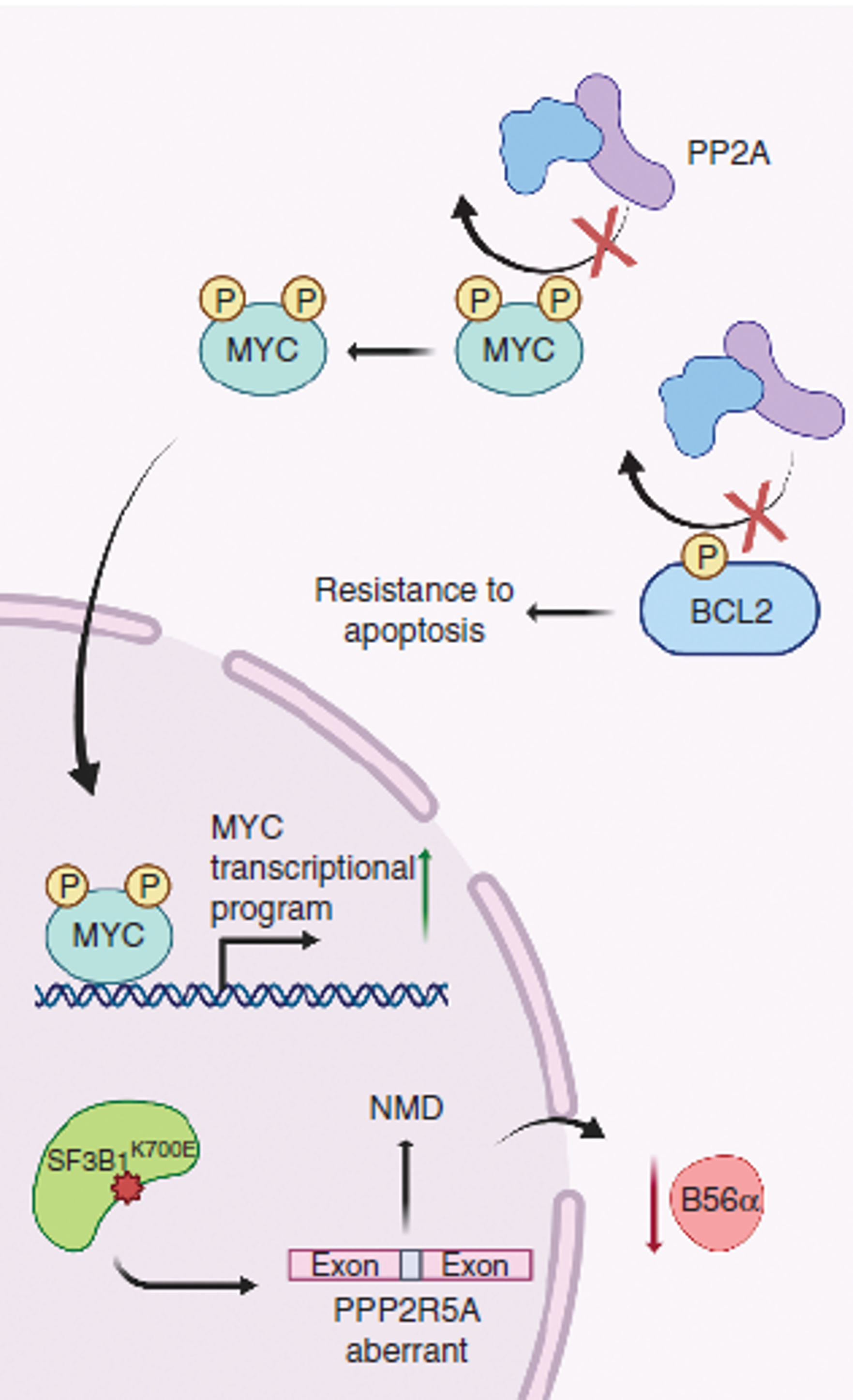
Aberrant RNA splicing
RNA splicing is a critical regulator of gene expression and mediator of proteome diversity. The most commonly reported spliceosomal mutations converged to genes including: SF3B1, U2AF1, SRSF2 and ZRSR2 (loss of function), which have been repeatedly identified in myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), chronic lymphocytic leukemia as well as other myeloid neoplasms. While alterations in splicing factor genes have been suggested to promote tumorigenesis, the discovery of recurrent, high frequency change-of-function mutations in RNA splicing factors (SF) in human cancers underscored a central role for altered splicing in hematologic malignancies and solid tumors. To study the effect of SF mutations on tumorigenesis, a few challenging steps are necessary: a) Rapid identification and accurate quantification of cancer-specific aberrant RNA splicing genome-widely, b) Determining the major changes of downstream cellular processes under splicing dysregulation in a systematic way, and c) Understanding the molecular mechanisms of tumorigenesis and fast translation to potential therapeutic strategy. All these steps require extensive computational efforts. Our lab will integrate knowledges in cancer genomics, systems biology, machine learning and computer science to address these challenging difficulties.
Precision Medicine
Outcomes of anticancer therapy vary dramatically among patients due to diverse genetic and molecular backgrounds, highlighting extensive intertumoral heterogeneity. The fundamental tenet of precision oncology defines molecular characterization of tumors to guide optimal patient-tailored therapy. Immunotherapies has proven to have improved objective response in tumors with high nonsynonymous mutation burden. T cell recognition of cancers relies upon presentation of tumor-specific antigens on MHC molecules, which generated by nonsynonymous mutations. However, recent studies suggested that tumor-specific alternative splicing events are far more abundant than the somatic single-nucleotide variants, and that peptides translated from intron-containing mRNAs are a unique class of neo-antigens in AML and other malignancies. In addition, spliceosome targeted therapies lead to global mis-processing of RNA and subsequent aberrant translation of intron encoded peptides. In this setting, we hypothesize that widespread accumulation of intron-encoded peptides due to RNA mis-splicing, would results in production of neo-antigens that can be targeted immunologically. We will collaborate with clinicians and pharmacists, and apply our expertise in computational biology to address this challenge.